Control Bar Features
The control bar at the top provides tools for data management, selection, export, and import.

1. Projection Selector
Switch between different dimensionality reduction methods:
| Method | Best For |
|---|---|
| PCA | Initial overview, finding outliers |
| UMAP | General exploration, balanced view |
| t-SNE | Finding clusters |
| PaCMAP | Fast alternative to t-SNE |
| MDS | Preserving distances |
Different projections reveal different patterns - try switching between them!
3D Projections
When a 3D projection is available, a plane selector (XY / XZ / YZ) appears, letting you view different 2D slices of the 3D space.
2. Annotation Selector
Choose which annotation to use for coloring points:
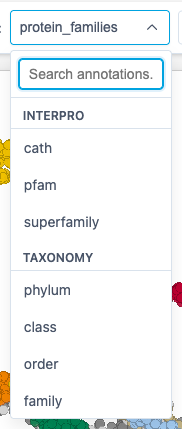
The Annotation dropdown features:
- Grouped categories: Features are organized into sections (UniProt, InterPro, Taxonomy, Other)
- Search: Type to filter features by name (case-insensitive)
- Keyboard navigation: Use arrow keys to navigate, Enter to select, Escape to close
Only categories present in your dataset appear in the dropdown. Any columns that don't match a known category appear under Other. See the ProtSpace Python package for the complete list of available annotations per source.
Tooltip-only annotations
gene_name, protein_name, and uniprot_kb_id are excluded from the dropdown but are still shown in the tooltip on hover.
3. Search
Find specific proteins by ID:
- Click inside the search box or press ⌘/Ctrl + K to focus it
- Type a protein ID or partial match
- Select from suggestions
- Protein is selected in the scatterplot
Multiple IDs
Paste multiple IDs at once (newline or space separated) and all matching proteins will be selected. Useful for re-selecting a previously exported subset.
4. Select Button
Enable box selection mode to select multiple proteins at once. See the Box Selection section for details.
5. Clear Button
Click Clear (or press Escape) to remove all current selections. Pressing Escape again will exit selection mode.
6. Isolate Button
Isolate focuses on selected proteins by hiding all others:
- Select one or more proteins (using search, click, or box select)
- Click Isolate
- Only selected proteins remain visible
- Click Reset (appears when isolated) to restore all proteins
Use Case
Isolate is useful for examining relationships within a specific protein subset - hiding unrelated proteins reduces visual clutter.
7. Filter Button
Filter shows only proteins matching specific annotation criteria:
- Click Filter to open the filter menu
- Select one or more annotations to filter by (e.g., taxonomy, family)
- Choose specific values for each annotation
- Click Apply
Filtered proteins are highlighted with a custom color scheme.
Isolate vs Filter
- Isolate: Works with selected proteins - hides everything else
- Filter: Works with annotation values - highlights matches, dims non-matches
Use Isolate for ad-hoc selections (like search results). Use Filter for annotation-based queries (like "show all kinases").
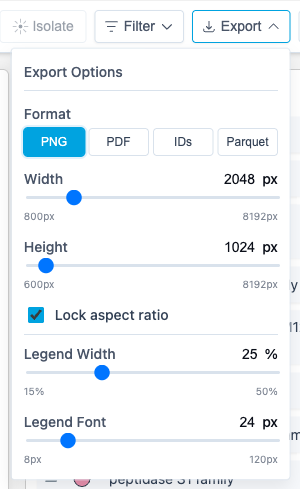
8. Export
Click Export to save your visualization:
| Format | Description |
|---|---|
| PNG | Raster image with legend |
| PDF document with legend | |
| Protein IDs | Text file with newline-separated identifiers |
| Parquet | .parquetbundle file with all data and optional settings |
See Exporting Results for image customization options (dimensions, legend size, font).
9. Import
Click Import to load a .parquetbundle file from your computer.
You can also drag & drop files directly onto the scatterplot.
Tips
- Compare projections: Patterns that appear in multiple projections are more reliable
- Use search: Quickly find known proteins to orient yourself
- Export often: Save interesting views for later
Next Steps
- Viewing 3D Structures - AlphaFold integration
- Exporting Results - Detailed export options